Barcelona, Spain (UroToday.com) A significant proportion of patients with metastatic prostate cancer have tumor cells harboring germline and/or somatic homologous recombination repair (HRR) gene alterations. Alterations in HRR genes such as BRCA1, BRCA2, and others are recognized to confer sensitivity to PARP inhibitors to varying extents. The PROfound study (NCT02987543) is the first positive Phase 3, randomized, multicenter clinical trial evaluating the efficacy and safety of the PARP inhibitor olaparib versus physician’s choice of enzalutamide of abiraterone in pre-treated metastatic castration-resistant prostate cancer (mCRPC) patients with a qualifying HRR gene alteration. To be eligible for the study, patients had to have documented tumor tissue qualifying mutations in one of 15 HRR genes that were predicted to be deleterious or suspected deleterious. Patients were screened from 20 countries including the USA, Europe, and Asia.
This abstract reports the prevalence of HRR single and co-occurring gene alterations derived from analysis of the 15-gene panel in patients screened for PROfound, including BRCA1, BRCA2, ATM, BRIP1, BARD1, CDK12, CHEK1, CHEK2, FANCL, PALB2, PPP2R2A, RAD51B, RAD51C, RAD51D, RAD54L.
Of the 4425 patients screened, 4047 had samples tested, of which only 2792 (69%) were successfully sequenced. Among the 31% of patients who failed screening, this was most commonly due to samples not meeting pathology requirements or insufficient yields of DNA after extraction, highlighting the importance of selecting high-quality samples to improve testing success rates.
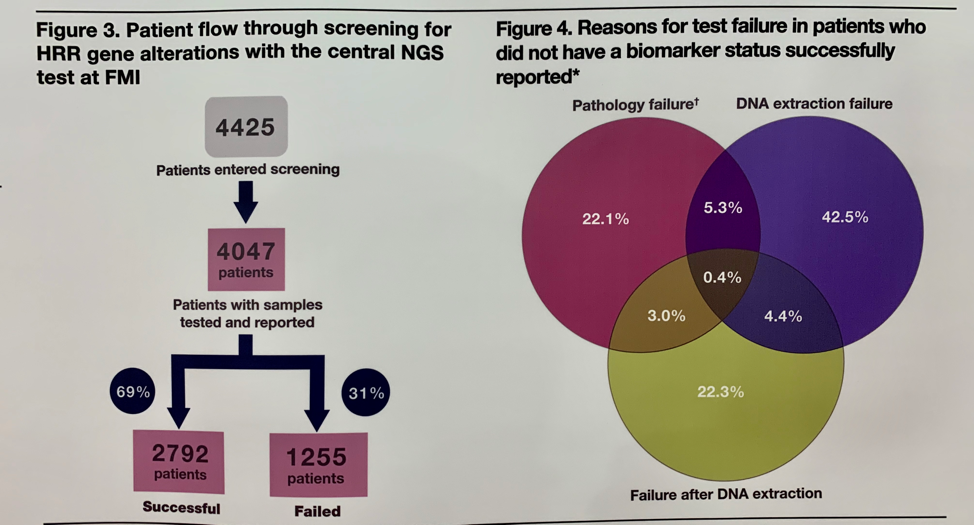
Most samples were from archived tissue (89.9%) and the majority of these were from the primary tumor (79.7%). A qualifying HRR alteration was detected in 27.9% of screened patients. A higher prevalence of HRR gene alterations was found in metastatic tumor than the primary tumor. The most common alterations were seen in BRCA2 (8.7%), CDK12 (6.3%), and ATM (5.9%). Co-occurring alterations were seen in 2.1% of sequenced tumors with the most commonly co-altered genes being BRCA2 and CDK12.
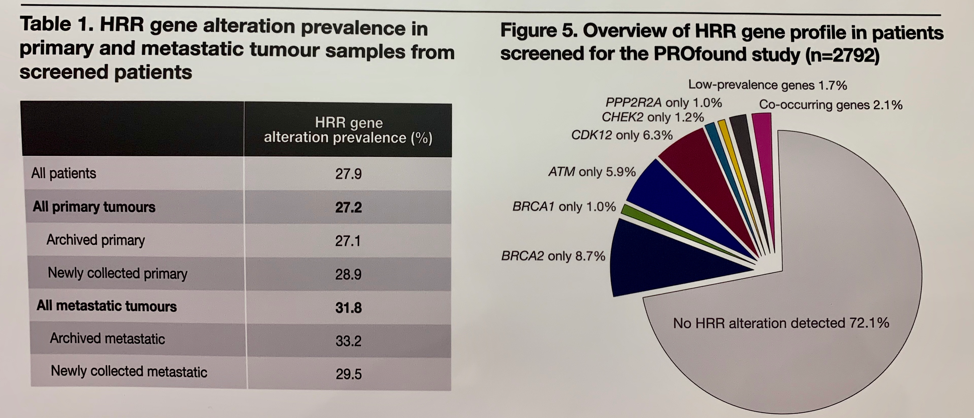
This study confirms that a significant number proportion of patients of mCRPC patients with alterations in HRR genes can be identified through tissue testing, though highlights the importance of further developing testing methodologies for analyzing different samples and biopsy types. The positive results of the PROfound study will be presented later at ESMO.
Presented by: Johann S. De Bono, MB, ChB, FRCP, MSc, PhD, FMedSci, Professor of Cancer Research and Professor in Experimental Cancer Medicine at The Institute of Cancer Research and The Royal Marsden NHS Foundation Trust, London, UK
Written by: Jacob Berchuck, MD, Medical Oncology Fellow at the Dana-Farber Cancer Institute (Twitter: @jberchuck) at the 2019 European Society for Medical Oncology annual meeting, ESMO 2019 #ESMO19, 27 Sept – 1 Oct 2019 in Barcelona, Spain
